When Kaggle finally launched a new tabular data competition after all this time, at first, everyone got excited. Until they weren’t. When the Kagglers found out that the dataset was 50 GB large, the community started discussing how to handle such large datasets [4].
CSV file format takes a long time to write and read large datasets and does not remember data types unless explicitly told.
Aside from reducing the required disk space by reducing the data types, the question was in which format to save the modified dataset in between work sessions [4]. The CSV file format takes a long time to write and read large datasets and also does not remember a column’s data type unless explicitly told. This article explores four alternatives to the CSV file format for handling large datasets: Pickle, Feather, Parquet, and HDF5. Additionally, we will look at these file formats with compression.
This article explores the alternative file formats with the pandas library. Now, you might be thinking “Why would you even use pandas when working with large datasets?” – That is a fair question. Although an alternative to pandas like Datatable would be faster at reading and writing CSV files, the advantage of pandas is that it offers a lot of flexibility for data processing. Additionally, pandas supports reading and writing a lot of file formats out of the box. Again, an alternative like Datatables does only support CSV, Jay, XLSX, and plain text formats [3].
You can find the code in my Kaggle Notebook.
Benchmark Setup
For the purpose of benchmarking, we will create a fictional dataset. This fictional dataset contains one column of each data type with the following exceptions: Columns with the data type float16 and categorical are omitted for this example because parquet does not support float16 and HDF5 with format = "table" does not support categorical. To reduce timing noise for comparability, this fictional dataset contains 10,000,000 rows and is almost 1GB large as suggested in [8].
)](images/how-to-handle-large-datasets-in-python-head-of-fictional-dataset-for-benchmarking-image.webp)
The characteristics of the data can impact reading and writing times, e.g. data types, width (number of columns) vs. length (number of rows) of DataFrame. However, this is beyond the scope of this article. For further reading, I recommend the following resources:
Overview of File Formats
In this section, we will briefly touch on some key characteristics for each file format: short description, file extension, used compression, and pandas reading and writing methods.
Comma-Separated Values (CSV)
A text file that uses a comma to separate values. The file extension is .csv.
In this article, we will use gzip compression.
# Reading
df = pd.read_csv(file_name,
dtype = {...})# Writing
df.to_csv(file_name,
index = False,
compression = ...) # None or "gzip"Pickle
The
picklemodule implements binary protocols for serializing and de-serializing a Python object structure. [7]
The file extension is .pkl.
In this article, we will use gzip compression.
# Reading
df = pd.read_pickle(file_name)# Writing
df.to_pickle(file_name,
compression = ...) # None or "gzip"Parquet
Apache Parquet is a columnar storage format available to any project in the Hadoop ecosystem, regardless of the choice of data processing framework, data model, or programming language. [2]
The file extension is .parquet.
In this article, we will use the pyarrow engine and gzip compression.
# Reading
df = pd.read_parquet(file_name)# Writing
df.to_parquet(file_name,
engine = "pyarrow",
compression = ...) # None or "gzip"Feather
Feather is a portable file format for storing Arrow tables or data frames (from languages like Python or R) that utilizes the Arrow IPC format internally. Feather was created early in the Arrow project as a proof of concept for fast, language-agnostic data frame storage for Python (pandas) and R. [1]
The file extension is .feather.
Since gzip compression is not available for the feather format, we will use zstd compression instead.
# Reading
df = pd.read_feather(file_name)# Writing
df.to_feather(file_name,
compression = ...) # None or "zstd"Hierarchical Data Format (HDF5)
HDF5 is a data model, library, and file format for storing and managing data. It supports an unlimited variety of data types, and is designed for flexible and efficient I/O and for high volume and complex data. HDF5 is portable and is extensible, allowing applications to evolve in their use of HDF5. [5]
The file extension is .h5.
HDF5 has two format options:
"fixed", which is fast at writing [6]"table", which is slower but offers “flexible operations like searching/selecting subsets of the data” [6]
For reading and writing HDF5 files you need to install tables.
Since gzip compression is not available for the feather format, we will use zlib compression instead.
# Reading
df = pd.read_hdf(file_name)# Writing
df.to_hdf(file_name,
key = "df",
format = ..., # "fixed" or "table"
complib = ..., # None or "zlib"
complevel = 9)Comparison
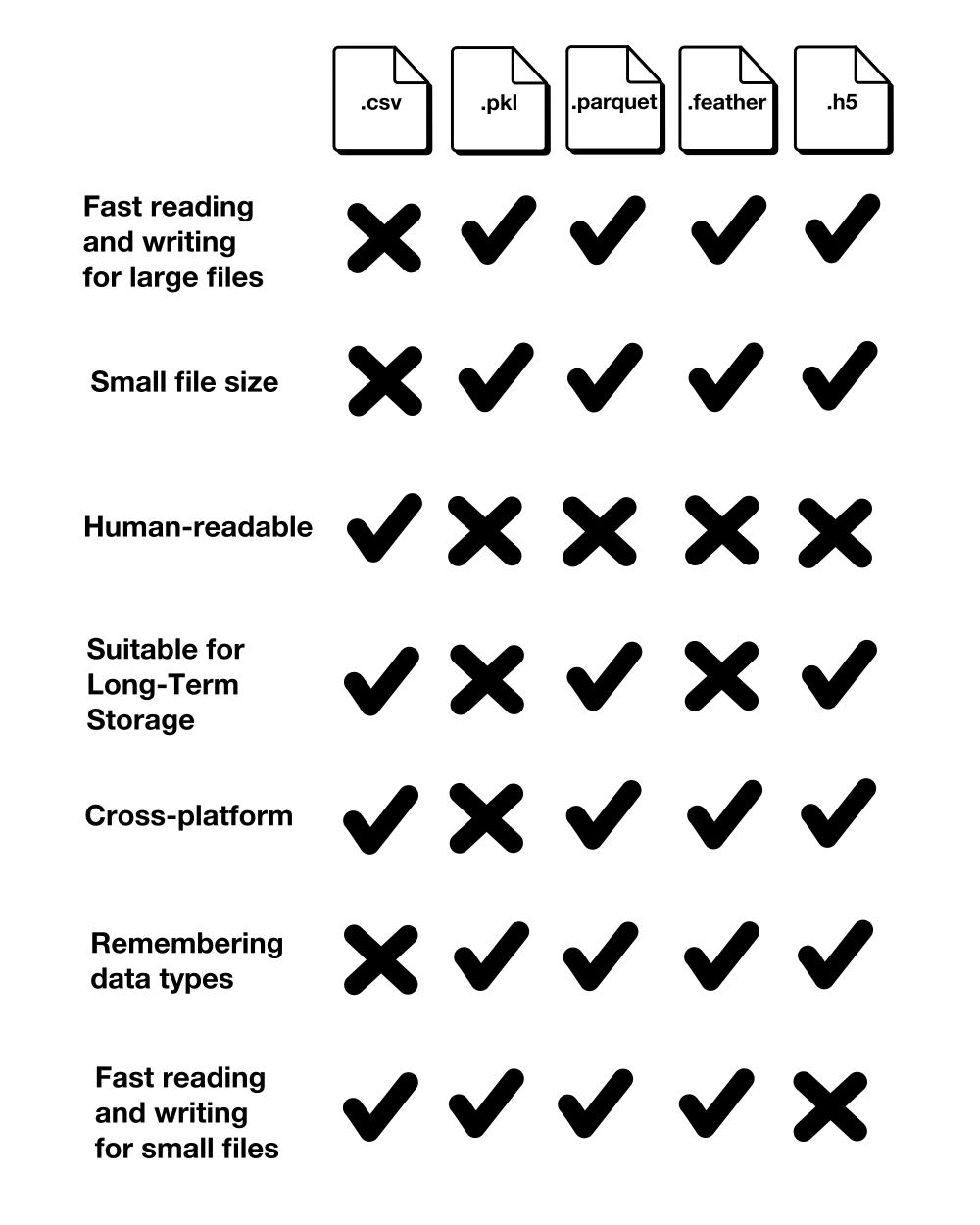
This section compares the five file formats according to writing time, file size, reading time, and miscellaneous characteristics such as human-readability, consistency, intended storage time and portability, and performance on small datasets.
Writing Times
Below, you can see the time it takes to write the file for each file format. The solid black bars indicate the writing times for the uncompressed files, while the hashed bars indicate the writing times for the compressed files.
)](images/how-to-handle-large-datasets-in-python-comparison-of-writing-times-for-different-file.webp)
First, let’s address the elephant in the room: Compression increases the writing time of any file format. But this should not be surprising as compression of data is an additional task during the writing process.
Additionally, we can make the following observations:
- As expected, for CSV the writing takes the longest
- Feather and Parquet have the fastest uncompressed writing times
- Feather has the fastest writing time for both uncompressed and compressed files
- As expected, HDF5 with
format = "fixed"is faster thanformat = "table", but with compression HDF5 withformat = "fixed"is similar toformat = "table"
File Sizes
Below, you can see the resulting file sizes for each file format. The solid black bars indicate the file sizes for the uncompressed files, while the hashed bars indicate the file sizes for the compressed files.
)](images/how-to-handle-large-datasets-in-python-comparison-of-file-sizes-for-different-file.webp)
We can make the following observations:
- As expected, the compressed files are smaller than the uncompressed files
- CSV is the largest file
- Parquet is the smallest uncompressed file
- Parquet and HDF5 with
format = "table"are the smallest compressed files
Reading Time
Below, you can see the time it takes to read the file for each file format. The solid black bars indicate the reading times for the uncompressed files, while the hashed bars indicate the reading times for the compressed files.
)](images/how-to-handle-large-datasets-in-python-comparison-of-reading-times-for-different-file.webp)
We can make the following observations:
- As expected, for CSV the reading takes the longes
- Pickle and HDF5 with
format = "fixed"have the fastest reading times for both uncompressed and compressed files - As expected, HDF5 with
format = "fixed"is faster thanformat = "table", also with compression
Miscellaneous
Aside from the reading and writing times and the file sizes, we should also look at a few more characteristics: Human-readability, consistency, intended storage time and portability, and performance on small datasets.
Human-Readability
CSV is the only file format that is readable by a human. All alternative file formats are binary formats and therefore not readable by humans.
Consistency
Unless you explicitly tell the .read_csv() method with which data type to read each column, the CSV file format does not remember the data types. This requires previous knowledge of the data types and is also an additional effort.
If the data types are not explicitly listed in the reading method, this can lead to increased required storage space as all integers are read as int64, all floats are read as float64, and datetime64[ns] and categorical are read as object. Below, you can see the original DataFrame and the DataFrame after writing to and reading from a CSV file:
)](images/how-to-handle-large-datasets-in-python-comparison-of-column-data-types-before-and-after.webp)
As you can see due to the casting of the smaller data types to larger data types, the required memory usage increases.
Intended Storage Time and Portability
Pickle and Feather are intended for short-term storage [7, 8, 9]. Pickle is intended for saving Python objects between work sessions and therefore is only supported by Python. Feather is intended for exchanging data between Python and R [9]. Both Pickle and Feather are also not guaranteed to be stable between versions [7, 9].
Performance on Small Datasets
Although, this article focuses on large datasets, it is noteworthy to mention the poor reading and writing times of HDF5 format for small datasets. As shown below, reading an HDF5 file takes even longer than a CSV file if the dataset is less than 2 MB.
)](images/how-to-handle-large-datasets-in-python-comparison-of-writing-and-reading-times-for-the.webp)
The following table summarizes this section’s comparison:
Conclusion
As expected, the CSV file format is both the slowest at reading and writing the file. Unless you are optimizing for file size, all alternatives are roughly only half of the CSV file format.
Unfortunately, the one-million-dollar question has an unsatisfying answer:
“Which is the best format to handle large datasets?”
– “It depends on your use case but probably not CSV”
Here is my attempt at proving a little more detail for the “It depends” part of this answer. I am interested to hear your input on this.
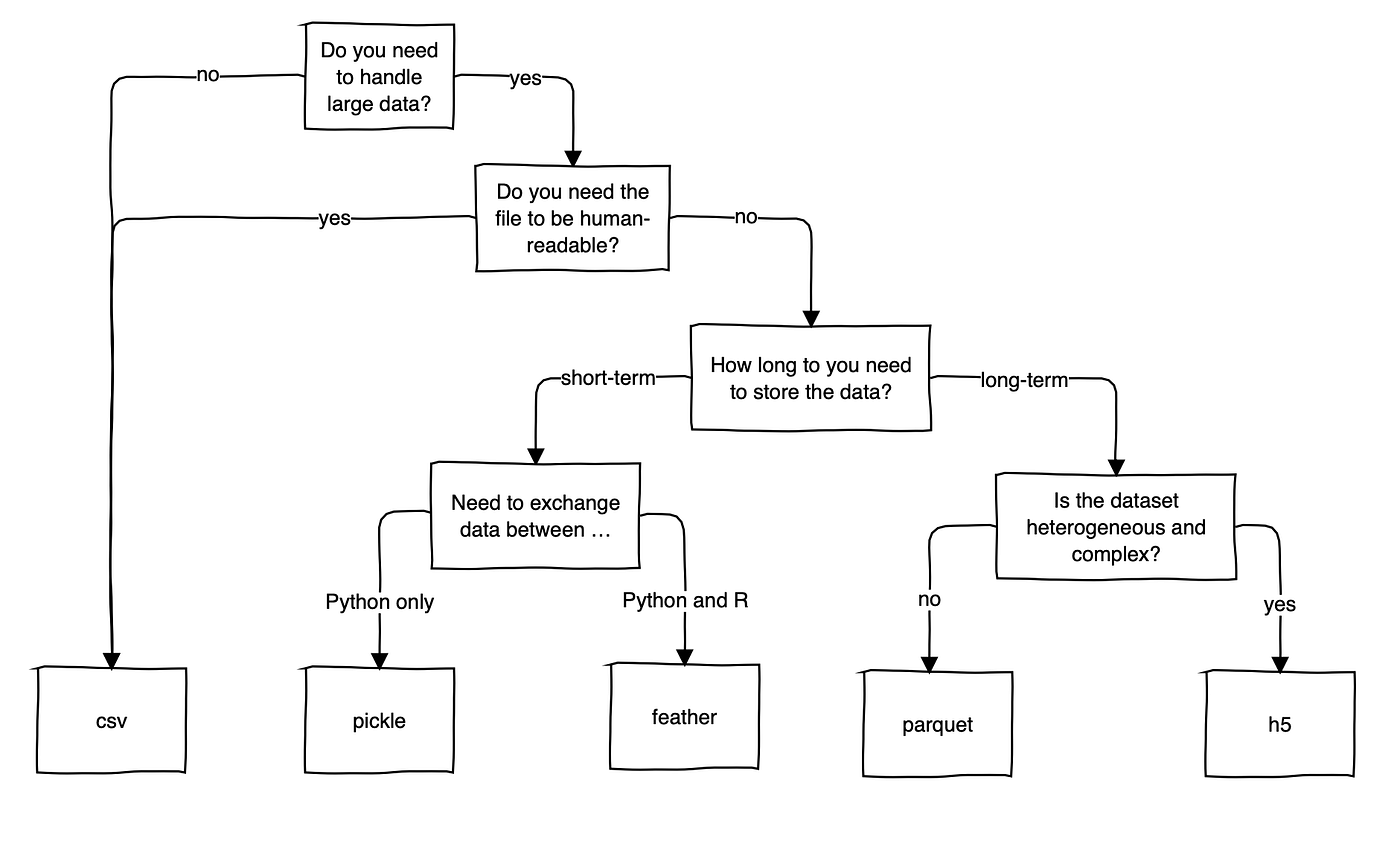
Despite its drawbacks, CSV is widely used. It seems like the fact that it is human-readable and not a binary file format makes it an intuitive file format, which you can quickly open up and have a look at without any trouble. So, unless you are working with multiple Gigabytes of data, CSV is still an acceptable option.
“CSV is not so bad after all. Leave it alone!”
References
[1] Apache Arrow, “Feather File Format”. apache.org. https://arrow.apache.org/docs/python/feather.html (accessed July 25, 2022)
[2] Apache Parquet, “Overview”. apache.org. https://parquet.apache.org/docs/overview/ (accessed July 25, 2022)
[3] Datatable, “datatable.fread()”. datatable.readthedocs.io. https://datatable.readthedocs.io/en/latest/api/dt/fread.html (accessed July 25, 2022)
[4] C. Deotte, ” How To Reduce Data Size”. kaggle.com. https://www.kaggle.com/competitions/amex-default-prediction/discussion/328054 (accessed July 25, 2022)
[5] The HDF Group, “HDF5”. hdfgroup.org. https://portal.hdfgroup.org/display/HDF5/HDF5 (accessed July 25, 2022)
[6] “pandas”, “pandas.DataFrame.to_hdf”. pydata.org. https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.to_hdf.html (accessed July 25, 2022)
[7] “Python”, “pickle – Python object serialization.” python.org. https://docs.python.org/3/library/pickle.html (accessed July 25, 2022)
[8] “Stackoverflow”, “What are the differences between feather and parquet?”. stackoverflow.com. https://stackoverflow.com/questions/48083405/what-are-the-differences-between-feather-and-parquet (accessed July 25, 2022)
[9] H. Wickham, “Feather: A Fast On-Disk Format for Data Frames for R and Python, powered by Apache Arrow”. rstudio.com. https://www.rstudio.com/blog/feather/ (accessed July 25, 2022)
This blog was originally published on Towards Data Science on Jul 26, 2022 and moved to this site on Feb 1, 2026.